Today I met with Sam and Steven a couple times to figure out what we’re going to discuss this Thursday with Pam. Steven and Sam have come up with three pools that we feel are a good place to start because unfortunately there’s not enough RNA to do the original pooling plan. This post details all the info that I currently have on the plan for our discussion on Thursday.
Links to spreadsheets:
RAW csv (mainsheet): 20180702-crab-sampling-file.csv
Excel spreadhseet (tabs for each pool): 20180702-crab-sampling-file.xls
New plan:
Start out with 3 pools:
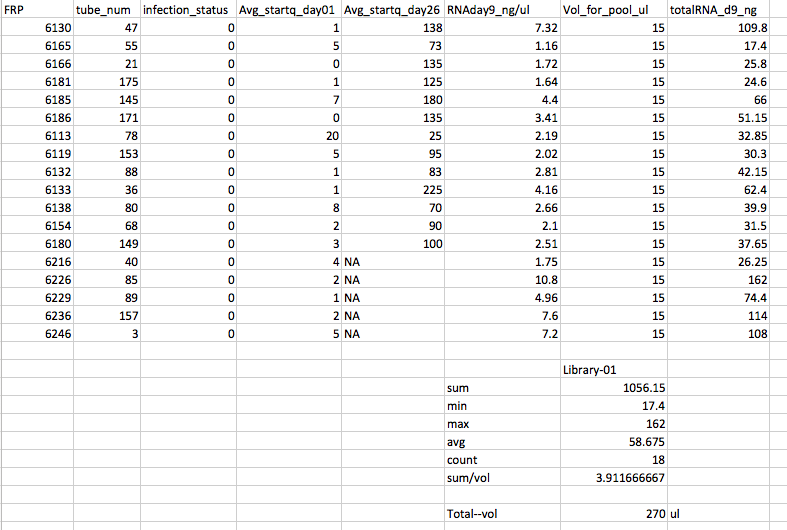
- Pool #1 Uninfected from Day 9
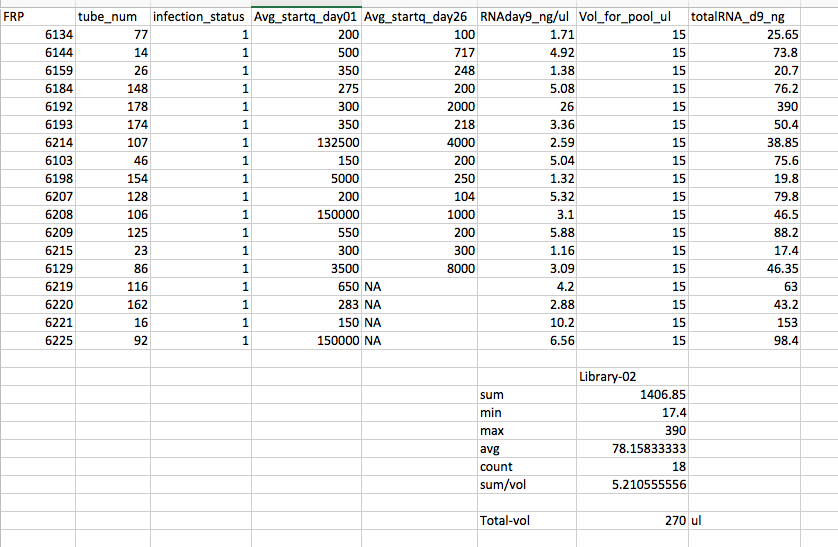
- Pool #2 Infected from Day 9
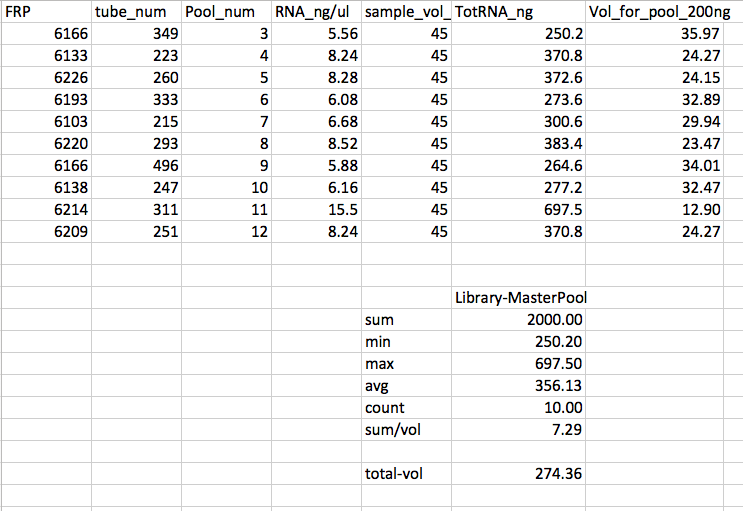
- MasterPool: The highest RNAng sample from each of the remaining 10 (Pools 3 - 12) pools
This plan will not be able to take temperature into account, but it will allow for gene discovery between the infected and uninfected (Pools 1 and 2). And gene discovery in the MasterPool.
After these pools are sequenced and some targets are ID’ed, we can go in and do qPCR and then plan for some more libraries based on those results.
Below are the details on how I am going to do the pools (very much subject to change as I get input from others):
Library #1 - Uninfected
Library #2 - Infected
MasterPool Library
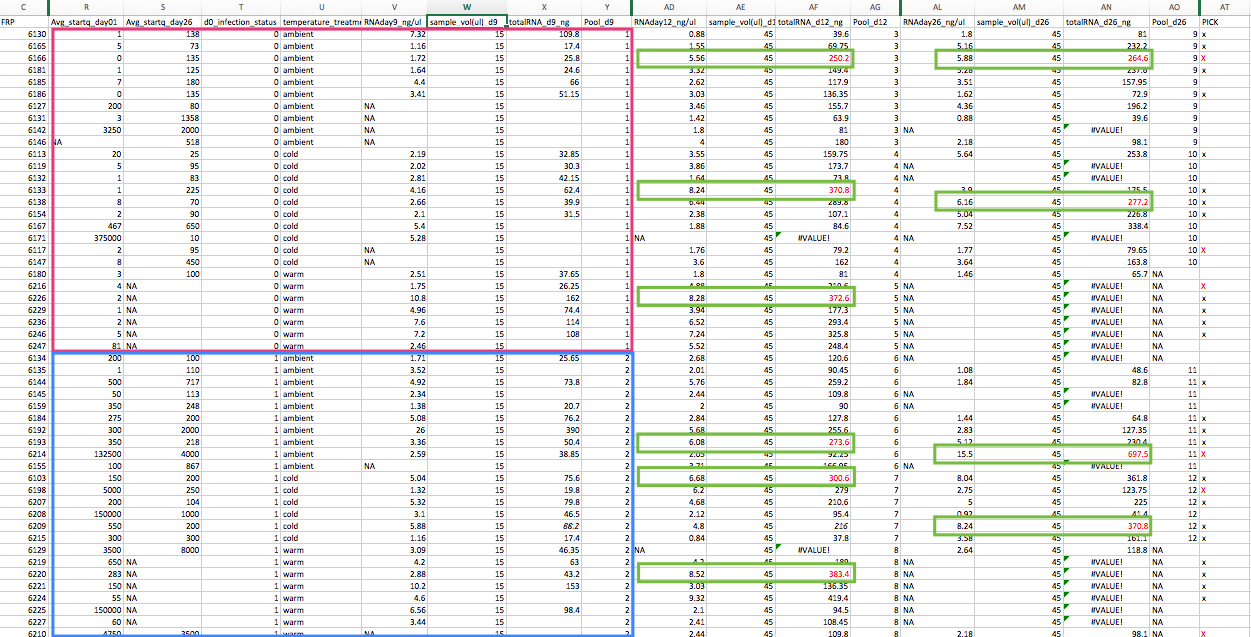
New Pool Plan in the big spreadsheet:
Pink: Library 1
Blue: Library 2
Green: MasterPool
Next Steps
If this all looks good, then we can move on to using the Speed Vac and then send them to get sequenced.