Today I played around with GitHub and R Projects. I’ve figured out a way of making adding Qubit results to my hemolymph and Qubit results master file more easy. It isn’t perfect and there are likely some things that I don’t know about that could make it easier, but I’ve detailed the new method below. I also noticed today that the tube_number column in what I did yesterday got all messed up with the merge in R. Detailed below.
New R Project and Script
Sam mentioned yesterday that it was difficult to help me because my R script and project was traversing two GitHub repositories.
So, today I worked on figuring out a way to make it easier not only for myself, but for future collaborations and help.
I made a new R Project and R script in the project-crab GitHub repository.
The script is much more straightforward and easier to use becuase the csv’s are already in this repository.
library(tidyverse)
# Script for creating joined and merged data files from project-crab. Ultimate goal is to create a true master file of all crab-related data files in this repo.
# Join all hemolymph sampling data with Qubit RNA HS results data into one file.
# read in all-crabs-hemo.csv
hemo <- read.csv("data/20180522-all-crabs-hemo.csv")
# read in Qubit-results.csv
qub <- read.csv("data/20180817-Qubit-results.csv")
# Convert "tube_number" in hemo.csv numeric (not factor)
hemo$tube_number <- as.numeric(hemo$tube_number)
# test that as.numeric conversion worked
is.numeric(hemo$tube_number)
# Join files based on column "tube_number"
master <- left_join(hemo, qub, by = "tube_number")
# Look at columns in master file
colnames(master)
# Select columns that are useful and relevant
master2 <- master %>%
select(year.FRP, FRP, infection_status, Uniq_ID, sample_day, infection_status, maturity, Holding_Tank, Low_Tag, High_Tag, sample, treatment_tank, where, tube_number, Comments, death, comments, CW, SC, CH, Test_Date, Original_sample_conc_ng.ul, total_sample_vol_ul, total_yield_ng) %>%
arrange(FRP)
# Write updated csv to project-crab data repo.
write.csv(master2, "20180817-hemo-Qubit.csv")
# Move new csv to data directory in repo project-crab and update readme.md
With this current process there are some things that I need to do outside of R. I add the new Qubit data to the 20180817-Qubit-results.csv file and add in the “total_sample_vol_ul” and the “total_yield_ng” values. Then, at the end of the script, I have to move the new csv to the data directory within the project-crab repository and update the readme.md.
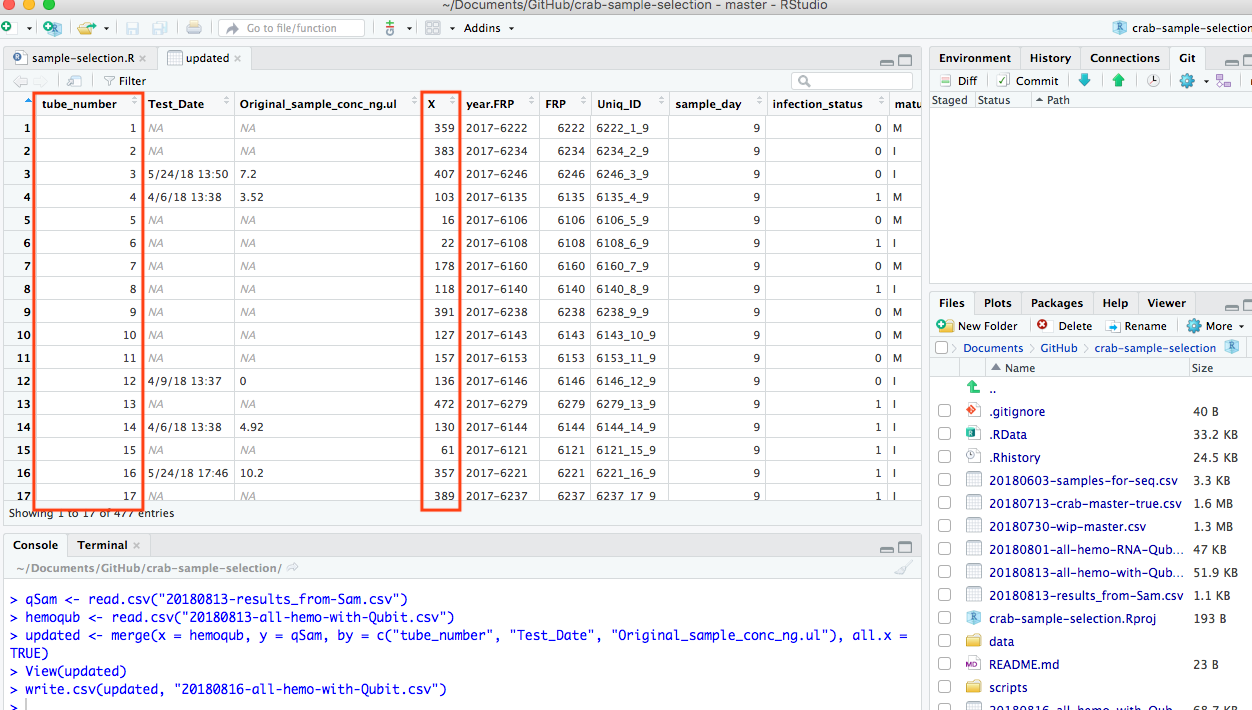
Merge in R messed up tube_number columns
This all lives in this repository: crab-sample-selection
R Script
R Project
```#Try to merge columns that are repeats. “Test_Date.x” with “Test_Date.y” and “Original_sample_conc_ng.ul.x” with “Original_sample_conc_ng.ul.y”.
qSam <- read.csv(“20180813-results_from-Sam.csv”) hemoqub <- read.csv(“../project-crab/data/20180813-all-hemo-with-Qubit.csv”)
updated <- merge(x = hemoqub, y = qSam, by = c(“tube_number”, “Test_Date”, “Original_sample_conc_ng.ul”), all.x = TRUE)
write.csv(updated, “20180816-all-hemo-with-Qubit.csv”) ```
Resulted in the new file’s columns getting messed up.
20180816-all-hemo-with-Qubit.csv
The tube numbers are in column marked “X” now…