More RNA isolation to replace those that had “out of range” RNA qubit results. Ended up contaminating them all, so unusable. Starting my journey of using R for data organization.
RNA Isolation
Today I process these samples:
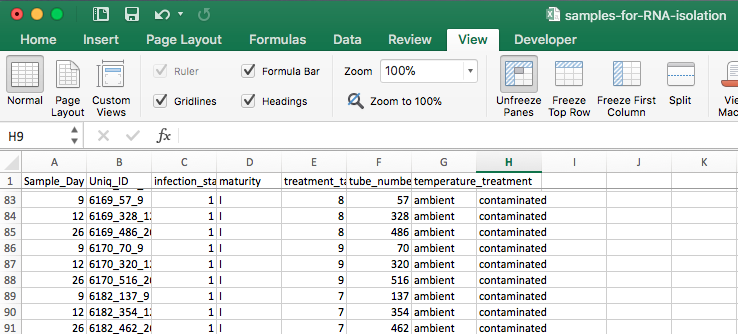
As you can see, there is a comment for each one saying “contaminated” because at the end of the isolation process when I was putting 50µL of 0.1% DEPC-treated water in each tube, I noticed by the fourth sample that the bottle with the water had a cloudy substance floating in it. I must have accidentally discarded the supernatant during the final alcohol wash into the bottle containing the water instead of the designated waste bottle.
The samples from the first sampling date for each of the three crabs was likely contaminated with the water, as such the remaining tubes are not usable either becuase they only encompass the second and third sampling dates for the crabs that I selected.
Huge bummer and now I know that I should always put the lid over the bottle even if it is just for a second so as to avoid making that mistake again. When you’re doing a protocol such as this that takes a long time and involves a lot of repetitive motion, you have to be super diligent.
I am going to be working from home tomorrow - organizing data sheets and editing and publishing the podcast - but since this weekend looks rainy, I’m going to come in to lab both days for a few hours at a time to get some more isolations done before the next crab meeting on Thursday!
Here is the set that I have chosen to replace the set that I messed up today:
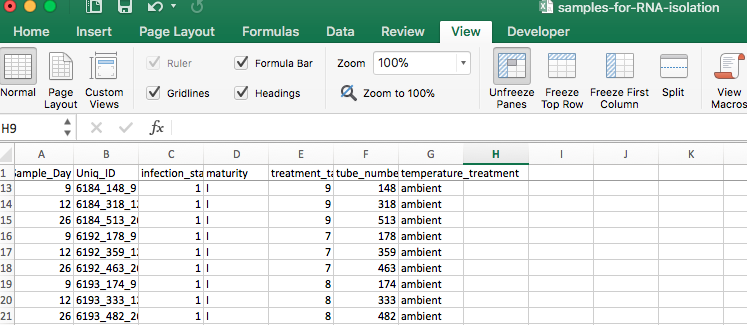
Luckily there are still a lot more samples to choose from!
Data organization
From Steven:
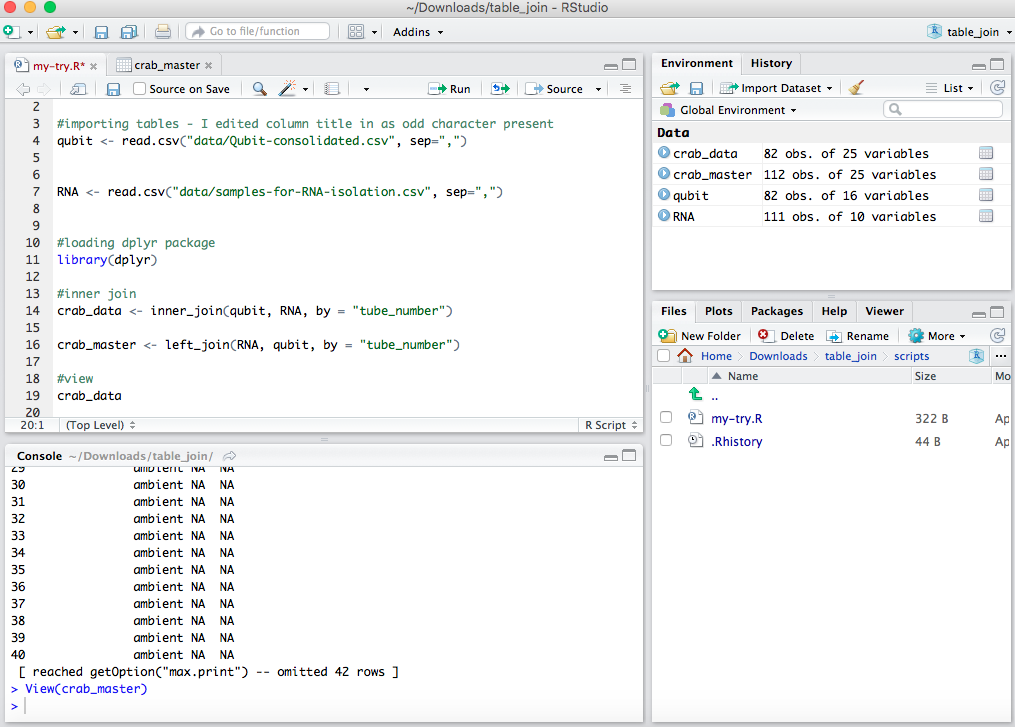
Spits out a file that joined the info based on the “tube_number” in common between the two spreadhseets. This was mostly done as a practice and tomorrow and over the weekend I’ll work more on my actual spreadsheets in GitHub crab project repo.