Today I BLASTn -ed Blue crab transcriptome with the assembled C. bairdi transcriptome on Mox. I also finished up some more things in my BLAST to GO GO-slim notebook with uniprot/sprot. Additionally, I got some input from Steven on how best to create a poster for GSS (which is this THURSDAY) so that I can get some cool stuff on there, but not use too many words… which is always a problem I have.
BLASTn
I ran blastn on Mox. (GitHub Issue #484)
First I made a database with the blue crab transcriptome:
make_blastn_db.ipynb
Then, I used this script to run blast on Mox:
#!/bin/bash
## Job Name
#SBATCH --job-name=1113_Cbairdi_blast_01
## Allocation Definition
#SBATCH --account=srlab
#SBATCH --partition=srlab
## Resources
## Nodes
#SBATCH --nodes=1
## Walltime (days-hours:minutes:seconds format)
#SBATCH --time=3-20:30:00
## Memory per node
#SBATCH --mem=100G
##turn on e-mail notification
#SBATCH --mail-type=ALL
#SBATCH --mail-user=graceac9@uw.edu
## Specify the working directory for this job
#SBATCH --workdir=/gscratch/srlab/graceac9/analyses/1113-Cb-blast
# Load Python Mox module for Python module availability
module load intel-python3_2017
/gscratch/srlab/programs/ncbi-blast-2.6.0+/bin/blastn \
-query /gscratch/srlab/graceac9/query/library01/query.fa \
-db /gscratch/srlab/graceac9/blastdb/bluecrab/blastdb/bluecrab \
-max_target_seqs 1 \
-outfmt 6 \
-num_threads 28 \
-out 1113-cbairdi-bc-blast.tab
I just got an email saying the job finished. I used nano to look at the file. Here’s some lines copy and pasted:
TRINITY_DN21432_c0_g1_i1 GEID01009651.1 79.268 246 26 17 $
TRINITY_DN21406_c0_g1_i2 GEID01112751.1 91.337 531 46 0 $
TRINITY_DN21488_c0_g1_i1 GEID01164208.1 100.000 31 0 0 $
TRINITY_DN21427_c0_g2_i3 GEID01037789.1 79.660 1293 236 22 $
TRINITY_DN21427_c0_g2_i2 GEID01037789.1 79.642 1174 212 22 $
TRINITY_DN21450_c0_g1_i1 GEID01013889.1 88.107 412 49 0 $
TRINITY_DN21468_c0_g2_i2 GEID01095198.1 96.429 56 2 0 $
TRINITY_DN5625_c2_g1_i3 GEID01080879.1 82.715 1886 310 16 410 $
TRINITY_DN5625_c2_g1_i4 GEID01080879.1 82.556 1886 313 16 410 $
TRINITY_DN5625_c2_g1_i5 GEID01080879.1 82.715 1886 310 16 378 $
TRINITY_DN5625_c2_g1_i2 GEID01080879.1 82.715 1886 310 16 410 $
TRINITY_DN5655_c0_g1_i1 GEID01030406.1 87.039 841 83 16 6 $
TRINITY_DN5655_c0_g2_i1 GEID01077734.1 94.900 451 14 5 1 $
TRINITY_DN5693_c0_g1_i1 GEID01052989.1 85.847 2805 362 15 454 $
TRINITY_DN5693_c0_g1_i3 GEID01052989.1 85.918 2805 363 14 454 $
TRINITY_DN5620_c0_g3_i1 GEID01164208.1 100.000 28 0 0 291 $
TRINITY_DN5670_c0_g2_i1 GEID01073871.1 82.257 1967 327 21 199 $
TRINITY_DN5672_c0_g1_i1 GEID01072687.1 89.773 88 7 2 1447 $
TRINITY_DN5634_c0_g1_i1 GEID01028809.1 86.521 549 52 16 180 $
TRINITY_DN5641_c1_g1_i1 GEID01073235.1 91.734 738 40 7 872 $
TRINITY_DN5666_c0_g4_i3 GEID01141318.1 83.922 1698 251 10 1 $
BLAST to GO GOslim
11052018-C_bairdi-blastn.ipynb
Finished up some things with that basing it on Steven’s notebook.
I took the final output from that file and moved it into my grace-Cbairdi-transcriptome/analyses directory.
I then used a new R script to make a weird pie chart.
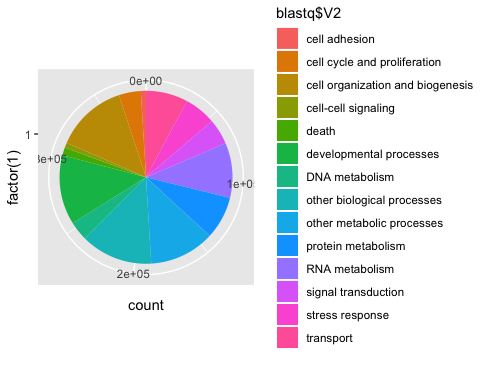
Because I’m trying to have something to put on a poster by tomorrow afternoon, I’ll just use excel, per Steven’s suggestion. Will learn the R or Jupyter notebook way once I don’t have a time crunch.
Trinity assembly stats using TrinityStats.pl
I tacked this on to the end of my notebook where I used Transrate.
Cbairdi_01_transrate.ipynb
Info on what the stats mean: here
Will use some of these stats on my poster.
################################
## Counts of transcripts, etc.
################################
Total trinity 'genes': 79739
Total trinity transcripts: 143172
Percent GC: 46.43
########################################
Stats based on ALL transcript contigs:
########################################
Contig N10: 3801
Contig N20: 2948
Contig N30: 2431
Contig N40: 2032
Contig N50: 1696
Median contig length: 608
Average contig: 1010.52
Total assembled bases: 144678598
#####################################################
## Stats based on ONLY LONGEST ISOFORM per 'GENE':
#####################################################
Contig N10: 3659
Contig N20: 2836
Contig N30: 2306
Contig N40: 1901
Contig N50: 1539
Median contig length: 479
Average contig: 873.95
Total assembled bases: 69687682
### Additional stuff done at home ## BLASTn with Hematodinium make_hemat_blastdb.ipynb
Upload blast db output to owl.
Run BLASTn on Mox.
Output in /gscratch/srlab/graceac9/analyses/1113-Cb-hemat-blastn.
Head of the output file 1113-cbairdi-hemat-blast.tab:
[graceac9@mox2 1113-Cb-hemat-blastn]$ head 1113-cbairdi-hemat-blast.tab
TRINITY_DN21452_c0_g1_i1 GEMP01003524.1 98.601 429 3 2 427 3862 4290 0.0 756
TRINITY_DN21452_c0_g2_i1 GEMP01003109.1 99.520 1874 8 1 154 2027 3975 2103 0.0 3410
TRINITY_DN21473_c0_g2_i1 GEMP01157246.1 94.161 137 1 1 137 240 369 8.40e-51 202
TRINITY_DN21473_c0_g1_i1 GEMP01004521.1 99.758 2478 5 1 119 2596 3699 1223 0.0 4542
TRINITY_DN21473_c0_g1_i1 GEMP01004521.1 95.833 72 3 0 53 124 3888 3817 2.70e-24 117
TRINITY_DN21498_c0_g2_i1 GEMP01004701.1 100.000 223 0 0 223 829 607 2.38e-114 412
TRINITY_DN21498_c0_g1_i1 GEMP01004701.1 98.523 474 7 0 17 490 629 156 0.0 837
TRINITY_DN21498_c0_g1_i1 GEMP01004701.1 100.000 46 0 0 53 608 563 1.38e-15 86.1
TRINITY_DN21405_c0_g1_i2 GEMP01169947.1 98.188 276 5 0 277 276 1 3.08e-135 483
TRINITY_DN21405_c0_g1_i1 GEMP01003188.1 99.445 541 3 0 542 541 1 0.0 983