Shipped off RNA for more sequencing yesterday! See post for details.
Samples Shipped:
| Sample_ID | species | treatment | experiment_day | bin_number | qubit_result_ng.uL | final_sample_vol | total_RNA_in_remaining_sample |
|---|---|---|---|---|---|---|---|
| PSC-421 | dermasterias_imbricata | exposed | 6 | 1 | 62.4 | 14 | 873.6 |
| PSC-422 | pisaster_ochraceus | exposed | 6 | 1 | 134 | 14 | 1876 |
| PSC-423 | pycnopodia_helianthoides | exposed | 6 | 1 | 42.4 | 14 | 593.6 |
| PSC-424 | dermasterias_imbricata | control | 6 | 9 | 16.2 | 14 | 226.8 |
| PSC-425 | pisaster_ochraceus | control | 6 | 9 | 154 | 14 | 2156 |
| PSC-426 | pycnopodia_helianthoides | control | 6 | 9 | 94.4 | 14 | 1321.6 |
| PSC-427 | dermasterias_imbricata | exposed | 6 | 2 | 30.2 | 14 | 422.8 |
| PSC-428 | pisaster_ochraceus | exposed | 6 | 2 | 124 | 14 | 1736 |
| PSC-429 | pycnopodia_helianthoides | exposed | 6 | 2 | 64.6 | 14 | 904.4 |
| PSC-430 | dermasterias_imbricata | control | 6 | 10 | 65.2 | 14 | 912.8 |
| PSC-431 | pisaster_ochraceus | control | 6 | 10 | 110 | 14 | 1540 |
| PSC-432 | pycnopodia_helianthoides | control | 6 | 10 | 130 | 14 | 1820 |
| PSC-433 | dermasterias_imbricata | exposed | 6 | 3 | 180 | 14 | 2520 |
| PSC-434 | pisaster_ochraceus | exposed | 6 | 3 | 72 | 14 | 1008 |
| PSC-435 | pycnopodia_helianthoides | exposed | 6 | 3 | 126 | 14 | 1764 |
| PSC-436 | dermasterias_imbricata | control | 6 | 11 | 86.6 | 14 | 1212.4 |
| PSC-437 | pisaster_ochraceus | control | 6 | 11 | 63.2 | 14 | 884.8 |
| PSC-438 | pycnopodia_helianthoides | control | 6 | 11 | 160 | 14 | 2240 |
| PSC-439 | dermasterias_imbricata | exposed | 6 | 4 | 19.4 | 14 | 271.6 |
| PSC-440 | pisaster_ochraceus | exposed | 6 | 4 | 92 | 14 | 1288 |
| PSC-441 | pycnopodia_helianthoides | exposed | 6 | 4 | TOO HIGH | 14 | #VALUE! |
| PSC-442 | dermasterias_imbricata | control | 6 | 12 | 164 | 12 | 1968 |
| PSC-443 | pisaster_ochraceus | control | 6 | 12 | 21.8 | 14 | 305.2 |
| PSC-444 | pycnopodia_helianthoides | control | 6 | 12 | 74 | 14 | 1036 |
| PSC-451 | dermasterias_imbricata | exposed | 6 | 6 | 144 | 14 | 2016 |
| PSC-452 | pisaster_ochraceus | exposed | 6 | 6 | 14.9 | 14 | 208.6 |
| PSC-453 | pycnopodia_helianthoides | exposed | 6 | 6 | 38.4 | 14 | 537.6 |
| PSC-454 | dermasterias_imbricata | control | 6 | 14 | 80.6 | 14 | 1128.4 |
| PSC-455 | pisaster_ochraceus | control | 6 | 14 | 130 | 14 | 1820 |
| PSC-456 | pycnopodia_helianthoides | control | 6 | 14 | 138 | 14 | 1932 |
| PSC-463 | dermasterias_imbricata | exposed | 6 | 8 | 148 | 14 | 2072 |
| PSC-464 | pisaster_ochraceus | exposed | 6 | 8 | 70.4 | 14 | 985.6 |
| PSC-465 | pycnopodia_helianthoides | exposed | 6 | 8 | 63 | 14 | 882 |
| PSC-466 | dermasterias_imbricata | control | 6 | 16 | 108 | 14 | 1512 |
| PSC-467 | pisaster_ochraceus | control | 6 | 16 | TOO HIGH | 14 | #VALUE! |
| PSC-468 | pycnopodia_helianthoides | control | 6 | 16 | 64.6 | 14 | 904.4 |
| PSC-520 | dermasterias_imbricata | control | 12 | 9 | 18.7 | 14 | 261.8 |
| PSC-521 | pisaster_ochraceus | control | 12 | 9 | 41.6 | 14 | 582.4 |
| PSC-522 | pycnopodia_helianthoides | control | 12 | 9 | 74.2 | 14 | 1038.8 |
| PSC-526 | dermasterias_imbricata | control | 12 | 10 | 97.8 | 14 | 1369.2 |
| PSC-527 | pisaster_ochraceus | control | 12 | 10 | 26.8 | 14 | 375.2 |
| PSC-528 | pycnopodia_helianthoides | control | 12 | 10 | 200 | 14 | 2800 |
| PSC-532 | dermasterias_imbricata | control | 12 | 11 | 55.2 | 14 | 772.8 |
| PSC-533 | pisaster_ochraceus | control | 12 | 11 | 33.8 | 14 | 473.2 |
| PSC-534 | pycnopodia_helianthoides | control | 12 | 11 | 126 | 14 | 1764 |
| PSC-538 | dermasterias_imbricata | control | 12 | 12 | 19.2 | 14 | 268.8 |
| PSC-539 | pisaster_ochraceus | control | 12 | 12 | 16.9 | 14 | 236.6 |
| PSC-540 | pycnopodia_helianthoides | control | 12 | 12 | 164 | 14 | 2296 |
| PSC-550 | dermasterias_imbricata | control | 12 | 14 | 26.6 | 14 | 372.4 |
| PSC-551 | pisaster_ochraceus | control | 12 | 14 | 27.4 | 14 | 383.6 |
| PSC-552 | pycnopodia_helianthoides | control | 12 | 14 | TOO HIGH | 14 | #VALUE! |
| PSC-562 | dermasterias_imbricata | control | 12 | 16 | 43.6 | 14 | 610.4 |
| PSC-563 | pisaster_ochraceus | control | 12 | 16 | 9.34 | 14 | 130.76 |
| PSC-564 | pycnopodia_helianthoides | control | 12 | 16 | 150 | 14 | 2100 |
So now, once I have the RNAseq data from these samples (23-28 business days), I’ll have the following for the multi-species project:
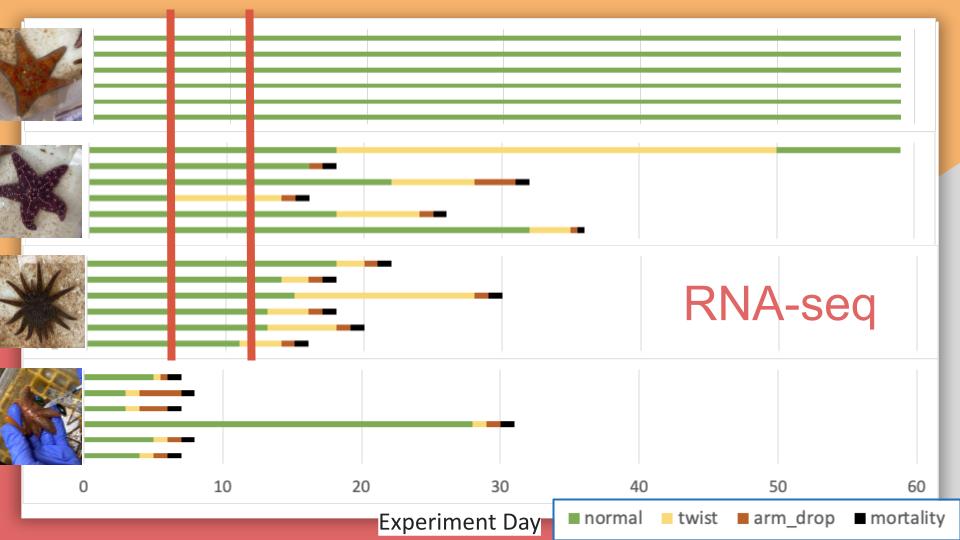
Controls and Exposed for Day 6 and Day 12
Note: Image only shows exposed stars disease signs. All controls had no disease signs.
Tracking Update
UPDATE - They received the samples!
I checked the tracking, and they signed for and received the samples :)