Building off of Wednesday’s post: Sep 18, 2024: Summer 2021 and Summer 2022 Analyses Status. Taking the DEG lists through enrichment (DAVID) and visualization (REVIGO).
DEG lists
Summer 2021 Annotated List:
paper-pycno-sswd-2021-2022/data/expB_DEG_DET_lists/DEGlist_8v8_2021_exposedVcontrol_counts_BLAST.tab
head -3 DEGlist_8v8_2021_exposedVcontrol_counts_BLAST.tab
transcriptID baseMean log2FoldChange lfcSE stat pvalue padj V2 uniprot_accession_ID gene V5 V6 V7 V8 V9 V10 V11 V12 V13 V14 PSC.56 PSC.52 PSC.54 PSC.61 PSC.64 PSC.73 PSC.76 PSC.81 PSC.59 PSC.57 PSC.69 PSC.67 PSC.71 PSC.75 PSC.78 PSC.83
g1746.t1 21.2778018566982 3.21964498222215 0.690679480502186 4.66156165502583 3.13819025114299e-06 3.3222665633176e-05 NA NA NA NA NA NA NA NA NA NA NA NA NA 3 2 2 4 9 10 10 3 5 11 8 30 13 26 12 77
g16359.t1 145.39071642552 -1.29337645882768 0.374317608050989 -3.45529152518921 0.000549698112215063 0.00274886552568119 sp Q8QGX4 PRKDC_CHICK 41.411 3557 1921 48 106 10569 347 3809 0 2622 371 223 334 161 191 231 355 411 60 30 34 312 87 7 126 30
Summer 2022 Annotated List:
paper-pycno-sswd-2021-2022/data/2022_DEG_DET_lists/DEGlist_2022_noage_contrast_counts_BLAST.tab
head -3 DEGlist_2022_noage_contrast_counts_BLAST.tab
transcriptID baseMean log2FoldChange lfcSE stat pvalue padj V2 uniprot_accession_ID gene V5 V6 V7 V8 V9 V10 V11 V12 V13 V14 PSC.0228 PSC.0187 PSC.0188 PSC.0174 PSC.0190 PSC.0231 PSC.0230 PSC.0219 PSC.0177 PSC.0186 PSC.0209 PSC.0203
g16359.t1 99.1727692502009 -0.907015523910786 0.317377350886998 -2.85784578318486 0.00426527631352983 0.016532402993508 sp Q8QGX4 PRKDC_CHICK 41.411 3557 1921 48 106 10569 347 3809 0 2622 43 81 83 46 29 72 291 92 83 193 166 173
g21290.t1 9.66291541993617 2.48401700624149 0.954169968115785 2.60332759282575 0.00923236895244479 0.0311189161798707 NA NA NA NA NA NA NA NA NA NA NA NA NA 60 13 6 0 6 8 13 4 7 0 0 3
DAVID
Background for both analyses is the list of uniprot accession IDs from the blast output of the genome gene list. paper-pycno-sswd-2021-2022/analyses/16-blast-annotation/blast_out_sep.tab
Summer 2021
GO_BP_DIRECT list:
paper-pycno-sswd-2021-2022/analyses/09-DAVID/2021_DAVID_output
Summer 2022
GO_BP_DIRECT list:
paper-pycno-sswd-2021-2022/analyses/09-DAVID/2022-DAVID_output_noage_contrast
REVIGO
Summer 2021
Input was the GO IDs from the 2021 DAVID output, and the values was the fold enrichment values for all terms that had a p-value less than 0.05.
Treemap:
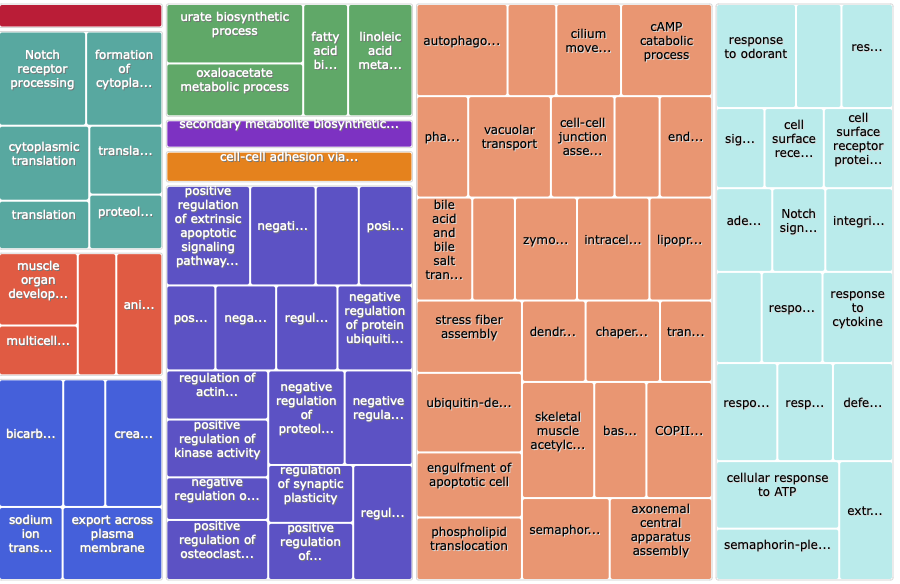
Treemap .tsv:
paper-pycno-sswd-2021-2022/analyses/revigo/2021_Revigo_BP_TreeMap.tsv
R code for the tree map downloaded from REVIGO:
paper-pycno-sswd-2021-2022/analyses/revigo/2021_Revigo_BP_TreeMap.R
Summer 2022
Input is the GO IDs from the 2022 DAVID output
Treemap:
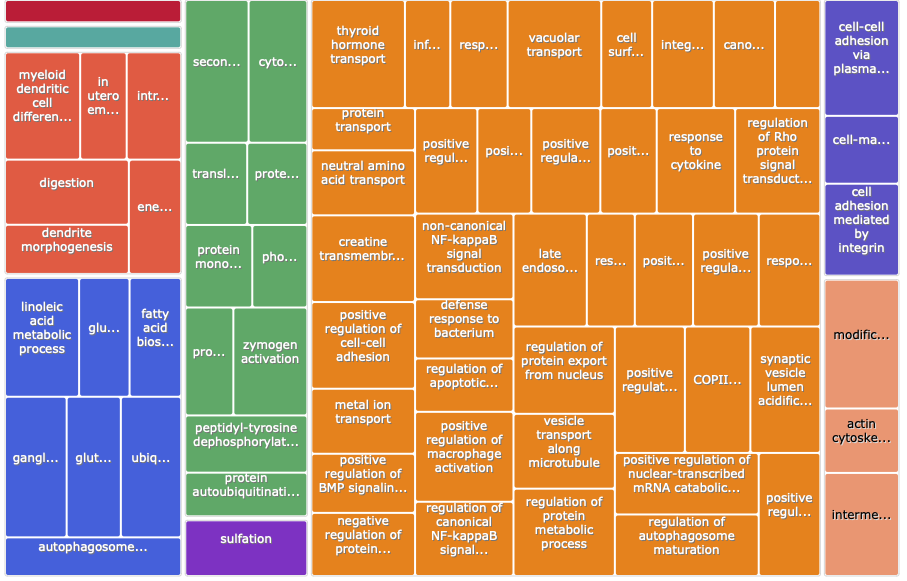
Treemap .tsv:
paper-pycno-sswd-2021-2022/analyses/revigo/2022_Revigo_BP_TreeMap.tsv
R code for the tree map downloaded from REVIGO:
paper-pycno-sswd-2021-2022/analyses/revigo/2022_Revigo_BP_TreeMap.R
Find out what DEGs are the same between the two datasets:
At the end of this Rmd: paper-pycno-sswd-2021-2022/code/18-deseq2_2021_2022.Rmd, I ran:
library(dplyr)
matchdegs <- inner_join(blastdeg2021counts, blastdeg2022counts, by = "transcriptID")
head(matchdegs)
4386 DEGs match between the two
DEG list of those that match between the two lists:
paper-pycno-sswd-2021-2022/data/DEGlist_2021-2022-matching.tab
There’s a ton of extra columns…. so it’s kinda messy.
DAVID
DAVID output of the 2021-2022 matching DEGs list with the genome gene list BLAST output uniprot accession IDs as the background: paper-pycno-sswd-2021-2022/analyses/09-DAVID/2021-2022-matched-DEGs_DAVID_output
REVIGO
Made with GO terms and Fold Enrichment values for all terms with p-value less than 0.05.
Treemap .tsv: paper-pycno-sswd-2021-2022/analyses/revigo/2021-2022-matchedDEGs_Revigo_BP_TreeMap.tsv
Treemap:
here
Get GO IDs for the DEGs
In uniprot.org tool for ID/Mapping, I pasted the uniprot accession IDs for the genome transcript list blast output. I added a column in the results for GO IDs, and downloaded the file to join and annotated the DEG lists. Downloaded as a .tsv: paper-pycno-sswd-2021-2022/data/genome_uniprot_GOIDs_2024_09_20.tsv
Summer 2021
Annotated DEG list: paper-pycno-sswd-2021-2022/data/expB_DEG_DET_lists/DEGlist_8v8_exVco_GOIDs.tab
Summer 2022
Annotated DEG list: paper-pycno-sswd-2021-2022/data/2022_DEG_DET_lists/DEGlist_2022_exVco_noagecontrast_GOID.tab