Post detailing the protocol for RNA extraction of the multispecies coelomocyte samples.
Sample Info
The samples are coelomocytes pelleted and preserved in 300ul of DNA/RNA Shield from Zymo Research and stored in -80C.
Here’s the protocol we used to sample and preserve the coelomocytes at Marrowstone - samples were later transferred to Roberts Lab -80 at SAFS:
Supply needs
- 29G 3/10cc, 1/2” insulin needles - for juvenile star collection
- Sterile, DNAse/RNAse-free microfuge tubes
- Labeled PSC ### for coelomocytes
- Labeled PSCS ### for coelomocyte supernatant
- Centrifuge
- DNA/RNA Shield
- P1000 and sterile tips
- Sample boxes - one for PSC samples, and one for PSCS samples
- -80C storage space
- Ice bucket with ice
- 70% ETOH spray bottle
- Paper towel
Protocol
- Collect coelomic fluid from the armpit using an insulin needle, wipe the needle with a paper towel sprayed with ETOH, and place in labeled microfuge tube
i. If need to do multiple sites, use a new needle every time - Collect as much volume as possible, with a max of 1mL (highly unlikely to collect this much)
- Place immediately on ice
- Bring to the dry lab
- Centrifuge for 5 minutes at 1200 rpm to separate the coelomocytes
- Remove the fluid using P1000 Pipette and tips and place in a labeled microfuge tube and store in -80C box for PSCS samples i. Note: will not be able to see a pellet, so act as though there is one at bottom of tube when removing supernatant
- Add 300 ul of DNA/RNA Shield to the pellet containing coelomocytes.
Store in -80C box for PSC samples
RNA Extraction
Kit: Zymo Research, Quick DNA/RNA Microprep Plus Kit (Cat D7005)
Extraction Preparations - labeling, etc
Let thaw:
- Proteinase K
- DNAse I
Label:
- 1 set of nuclease-free snap cap tube for each sample
- 1 set of Zymo-Spin IC-XM Column (yellow) plus the collection tube
- 1 set of Zymo=Spin IC Coolumn (clear) plus collection tube
- 1 nuclease-free tube for DNAse treatment
- 1 set of final labelled nuclease-free tube for eluted RNA.
I. Sample Preparation
Get samples from -80, and let thaw on wet ice, Mix well by vortex.
- Add 15ul Proteinase K and 30ul PK Digestion Buffer to each sample.
- Pipet to mix and let thaw at room temp for 30mins or longer.
- Vortex the sample after incubation and centrifuge at max speed for 2 minutes to pellet debris. Transfer 300ul of the cleared supernatant to a new nuclease-free tube (labeled).
- Add a 1:1 ratio (300ul) of DNA/RNA Lysis Buffer to the supernatant and mix well by pipetting.
II. DNA/RNA Purification
Perform all steps at 16,000g for 30sec unless specified.
- Transfer the sample (~600ul) to a Zymo-Spin IC-XM Column (yellow) in a collection tube and centrifuge at 16,000g for 30sec. i. FLOW-THROUGH HAS RNA!!! FILTER HAS DNA –> PUT IN -20C ii. Put DNA filter into a new collection tube and place in -20C freezer in FTR 213
- Add 1 volume (600ul) of 95-100% EtOH to the flow-through and mix by pipetting. i. Transfer the sample into a Zymo-Spin IC Column (clear) with collection tube ii. Discard flow-through
- DNase I Treatment
i. Add 400ul DNA/RNA Wash Buffer to the column and centrifuge at 16,000g for 30 sec. Discard flow-through
ii. Prepare DNase I Reaction Mix in labelled nuclease-free tube a. 5ul of DNase I : 35ul of DNA Digestion Buffer x ___ reps b. invert gently to mix iii. Add 40ul DNase I Reaction Mix to each sample and let incubate at room temp for 15 minutes - Add 400ul DNA/RNA Prep Buffer and centrifuge at 16,000g 30s. Discard flow-through
- Add 700ul DNA/RNA Wash Buffer and centrifuge at 16,000g 30s. Discard flow-through
- Add 400ul DNA/RNA Wash Buffer and centrifuge at 16,000g for 2 minutes. Discard flow-through and transfer column to a new, labelled nuclease-free tube.
- Add 15ul Room Temp DNAse/RNAse-Free Water directly to column and centrifuge 16,000g 30s.
Can now store at -80C or immediately run 1ul on Qubit.
Quantify RNA (Qubit)
Example ratios if had 4 samples to run:
4 samples, 2 standards, 1 extra prep = 4+2+1= 7 preps
Mix the Working solution:
7 x 1ul RNA HS Dye = 7 ul RNA HS dye
7 x 199ul RNA HS Buffer = 1393 ul RNA HS Buffer
Standards:
10ul S1 + 190ul working solution
10ul S2 + 190ul working solution
Samples:
1ul of sample + 199ul working solution.
Vortex well 5 sec.
Incubate RT 2 mins.
Run on Qubit with RNA HS, record data
Extraction Plan/Schedule
I currently have RNAseq data for Day 12 from 6 exposed bins. Each bin has 3 RNAseq libraries associated with it, one for P. helianthoides, one for P. ochraceus, and one for D. imbricata.
See figure showing the timepoint (red line) where current data is from:
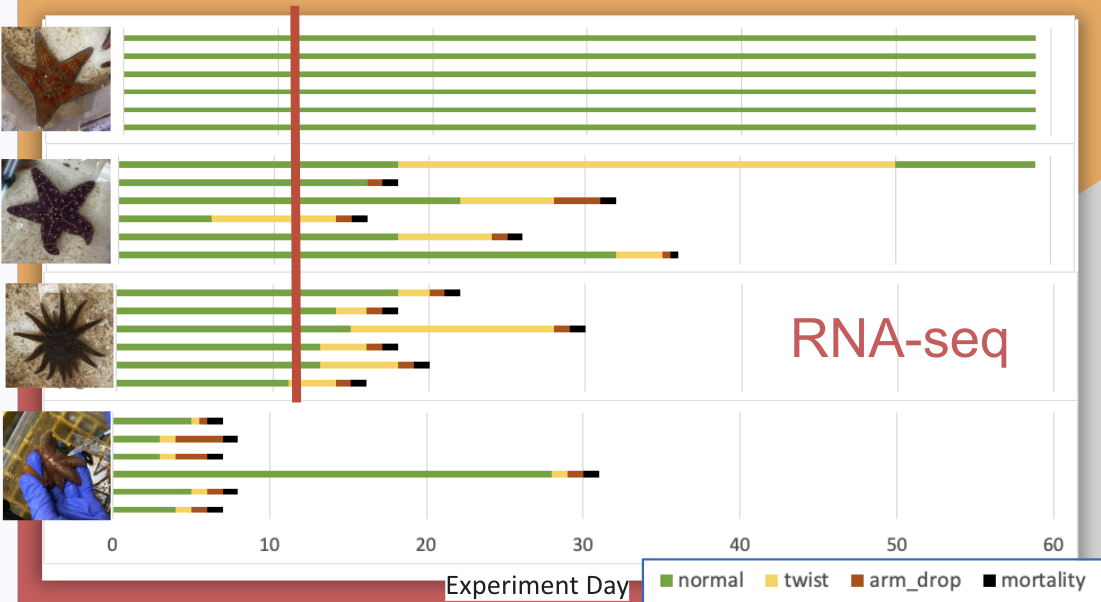
I have funding to send out 54 samples, with the following breakdown for the multi-species project:
Day 12 Controls
n=18 libraries
n=6 P. helianthoides
n=6 P. ochraceus
n=6 D. imbricata
Day 6 Exposed
n=18 libraries
n=6 P. helianthoides
n=6 P. ochraceus
n=6 D. imbricata
Day 6 Controls
n=18 libraries
n=6 P. helianthoides
n=6 P. ochraceus
n=6 D. imbricata
Total libraries = 18x3 = 54 libraries.
Extraction plan
Notes:
- I’m working remotely June 18-24 for a wedding and family time during which I’ll work on Day 12 exposed Multi-species data
- RNA samples need to be shipped early in the week to ensure that there’s no shipping issues to New Jersey… so I am aiming to send them out June 30th, July 1st, or July 2nd
Additionally, I’ll aim to extract in sets of 18, with one round of 4 samples to review the extraction before I jump into the main samples.
Schedule:
Note: I have one full box of 50 preps, plus 32 extra preps, so 82 total preps.
Tuesday, June 24th (I land at 2pm)–> afternoon:
- Do a practice extraction with 4 samples
- Run 1ul on Qubit
Wednesday, June 25th:
- Extract RNA from 18 samples
- Run 1ul on Qubit
Thursday, June 26th:
- Extract RNA from 18 samples
- Run 1ul on Qubit
Friday, June 27th:
- Extract RNA from 18 samples
- Run 1ul on Qubit
NOTE: Randomize the 54 samples across the three days so I’m not extracting RNA from each group.
Monday, June 30th:
- Prepare samples for shipping
If I can’t finish the extractions in time for June 30th, I can shift a little and do extraction of 18 samples on Monday June 30th and/or Tuesday July 1st. With shipping Tuesday July 1 or Wednesday July 2.