Today I picked samples for sending off to get sequenced. Obviously I have to get this checked with everyone… and I’m guessing they’d rather see it in a spreadhseet form and not color-coded as I do now… have to figure out how to do this.
Sample selection
This is probably super obnoxious and I’m sure a clear spreadsheet from using R is prefered, but here is a screenshot of the samples I picked for sequencing based on the master sheet that Steven made with all the RNA Qubit results and qPCR starting quantity results:
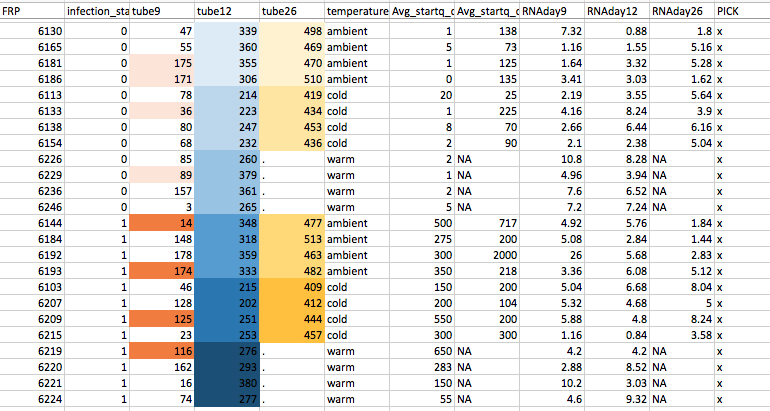
This is based on this .csv I made just pulling out the important columns…
The different colors are the different pools. There are 12 pools:
Day 9
(1) Uninfected
(2) Infected
Day 12
(3) Ambient, uninfected
(4) Cold, uninfected
(5) Warm, uninfected
(6) Ambient, infected
(7) Cold, infected
(8) Warm, infected
Day 26
(9) Ambient, uninfected
(10) Cold, uninfected
(11) Ambient, infected
(12) Cold, infected
The Day 12 and 26 pools were easy to pick.
The Day 9 pools are based on the Avg_startq_day01 column. The uninfected samples were selected based on the lowest starting quantity, and happened to be crabs that ended up in the three different temperature treatments. The same was done with the infected Day 9 samples, except that I selected based on the highest starting quantity.