Today I checked on my finished BLAST on Mox with my assembled C. bairdi transcriptome and the nt taxonomy database. It finished after three-ish days, and the file was large. However, all of the taxonomy cells were “N/A”. I made a GitHub Issue, and got some input and am now BLAST-ing again. Additionally, Sam posted a link for me to look into to get Order, Family, etc. taxonomy information on my next BLAST that I will get going before I leave for CA.
nt taxonomy BLAST: missing info
GitHub Issue 521
My output .tab file looked like this:
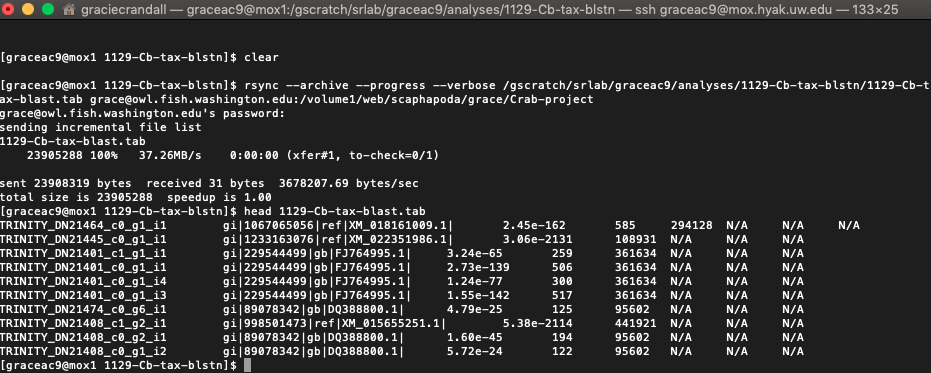
And another look in R (the script in the image was not saved as it is a copy of my original script):
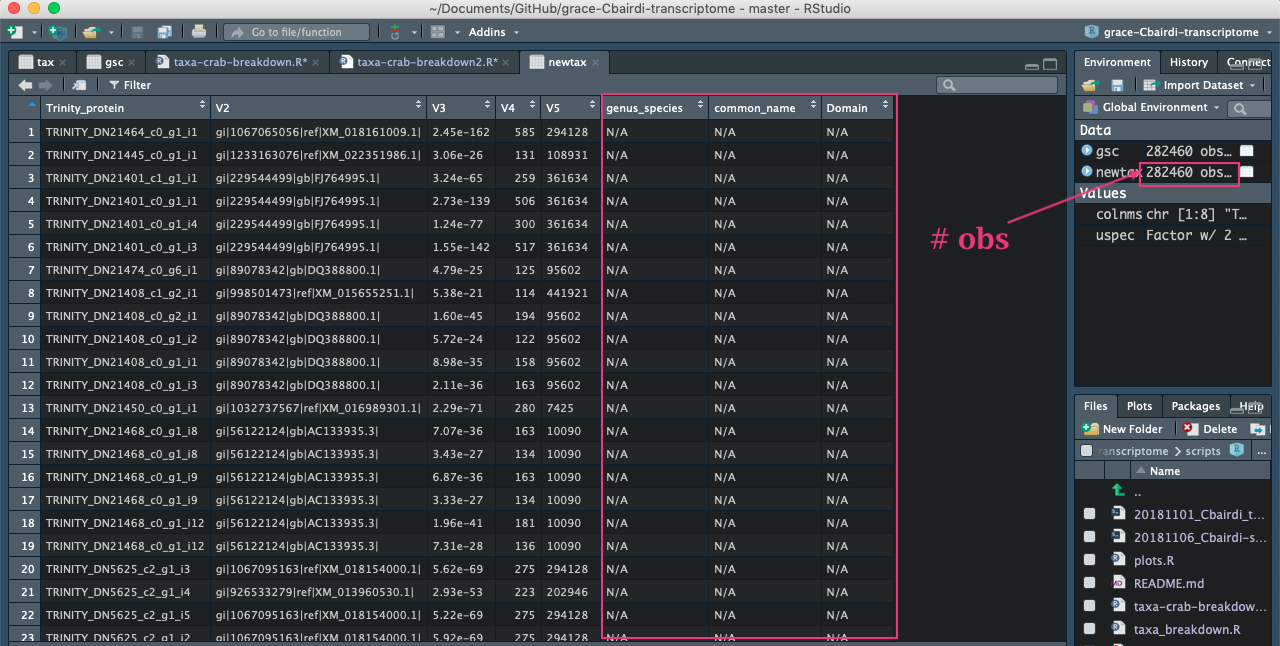
I changed the export command to export BLASTDB=/gscratch/srlab/blastdbs/ncbi-nr-nt-20181114/, because this is where the nt and taxonomy databases exist.
Re-running BLAST now, and it will be done in a couple of days.
BLAST nt taxonomy with Order tax infor
GitHub Issue 513
Currently, the output format is: -outfmt "6 qseqid sseqid evalue bitscore staxids sscinames scomnames sskingdoms" \, but I want to add more specific information like Order (DecaPod) and maybe Family.
URL to where I can find the information to do this as provided by Sam: ftp://ftp.ncbi.nih.gov/pub/taxonomy/taxdump_readme.txt
Will work on this this week so that it will be running before I get on my plane Friday.