Isolate RNA from warm crabs day 9 and 12 (crabs didn’t make it to day 26). Work more on creating a master file of all data associated with the crab project.
RNA isolation
Because only three crabs exposed to the warm temperature treatmeent survived the experiment, we decided during out meeting on Tuesday (2018-05-22) that I would isolate RNA from warm treatment crabs (infected and uninfected) that made it to the temperature treatment stage (day 12). So today I started that process and isolated RNA from 8 samples (4 crabs, all negative for Hematodinium based on Pam’s qPCR results).
Hemolymph collection data of samples I processed today:
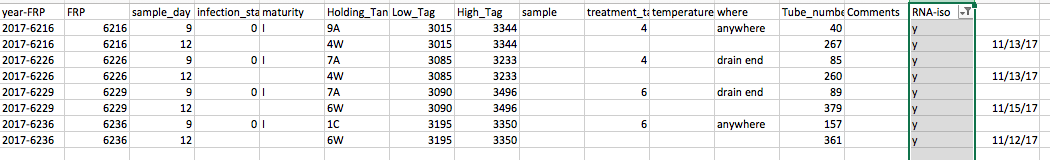
RNA HS Qubit results from those samples (tube number ties back to FRP in the hemolymph data sheet):
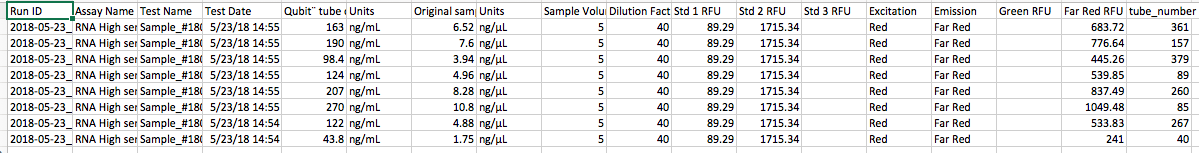
Tomorrow I will isolate RNA from two more crabs that are warm temperature treatment and uninfected (6 total uninfected) and 6 crabs that are warm treatment infected (using qPCR data from Pam).
Data organization
Working on figuring out a way to use R to organize the data into a master file such that each row is an individual crab (as designated by the unique FRP ID number), with columns for each piece of important data we have associated with that crab (hemolymph tube numbers from the sample dates, RNA isoaltion and Qubit data, morphology data, qPCR results). I am enjoying the puzzle-solving-like aspect of this process, but it can be overwhelming sometimes becuase it is a lot of data that is in many different sheets and workbooks. Our master file will include ALL crabs, including those that died after the second sampling date.
Thursday and Friday goals:
Thursday
- finish isolating RNA
- data organization and master file creation
Friday
- data organization and master file creation
- work on and finish a pooling scheme for RNA sequencing using the master file